Research
Microbiome
The recent realization that human-associated microbial communities play a crucial role in determining our health and well-being has led to the ongoing development of microbiome-based therapies such as fecal microbiota transplantation. Those microbial communities are very complex, dynamic and highly personalized ecosystems, exhibiting a high degree of inter-individual variability in both species assemblages and abundance profiles. It is not known whether the underlying ecological dynamics, which can be parameterized by growth rates, intra- and inter-species interactions in population dynamics models, are largely host-independent (i.e. “universal”) or host-specific. If the inter-individual variability reflects host-specific dynamics due to differences in host lifestyle, physiology, or genetics, then generic microbiome manipulations may have unintended consequences, rendering them ineffectual or even detrimental. Alternatively, microbial ecosystems of different subjects may follow a universal dynamics with the inter-individual variability mainly stemming from differences in the sets of colonizing species. Here we developed a novel computational method to characterize human microbial dynamics. Applying this method to cross-sectional data from two large-scale metagenomic studies, the Human Microbiome Project and the Student Microbiome Project, we found that both gut and mouth microbiomes display pronounced universal dynamics, whereas communities associated with certain skin sites are likely shaped by differences in the host environment. Interestingly, the universality of gut microbial dynamics is not observed in subjects with recurrent Clostridium difficile infection but is observed in the same set of subjects after fecal microbiota transplantation. These results fundamentally improve our understanding of forces and processes shaping human microbial ecosystems, paving the way to design general microbiome-based therapies.
Gene Regulation
A long-standing model holds that stochastic aberrations of transcriptional regulation play a key role in the process of ageing. While transcriptional dysregulation is observed in many cell types in the form of increased cell-to-cell variability, its generality to all cell types remains doubted. Here, we propose a new approach for analysing transcriptional regulation in single-cell RNA sequencing data by focusing on the global coordination between the genes rather than the variability of individual genes or correlations between pairs of genes. Consistently, across very different organisms and cell types, we find a decrease in the gene-to-gene transcriptional coordination in ageing cells. In addition, we find that loss of gene-to-gene transcriptional coordination is associated with high mutational load of a specific, age-related signature and with radiation-induced DNA damage. These observations suggest a general, potentially universal, stochastic attribute of transcriptional dysregulation in ageing.
Network Science
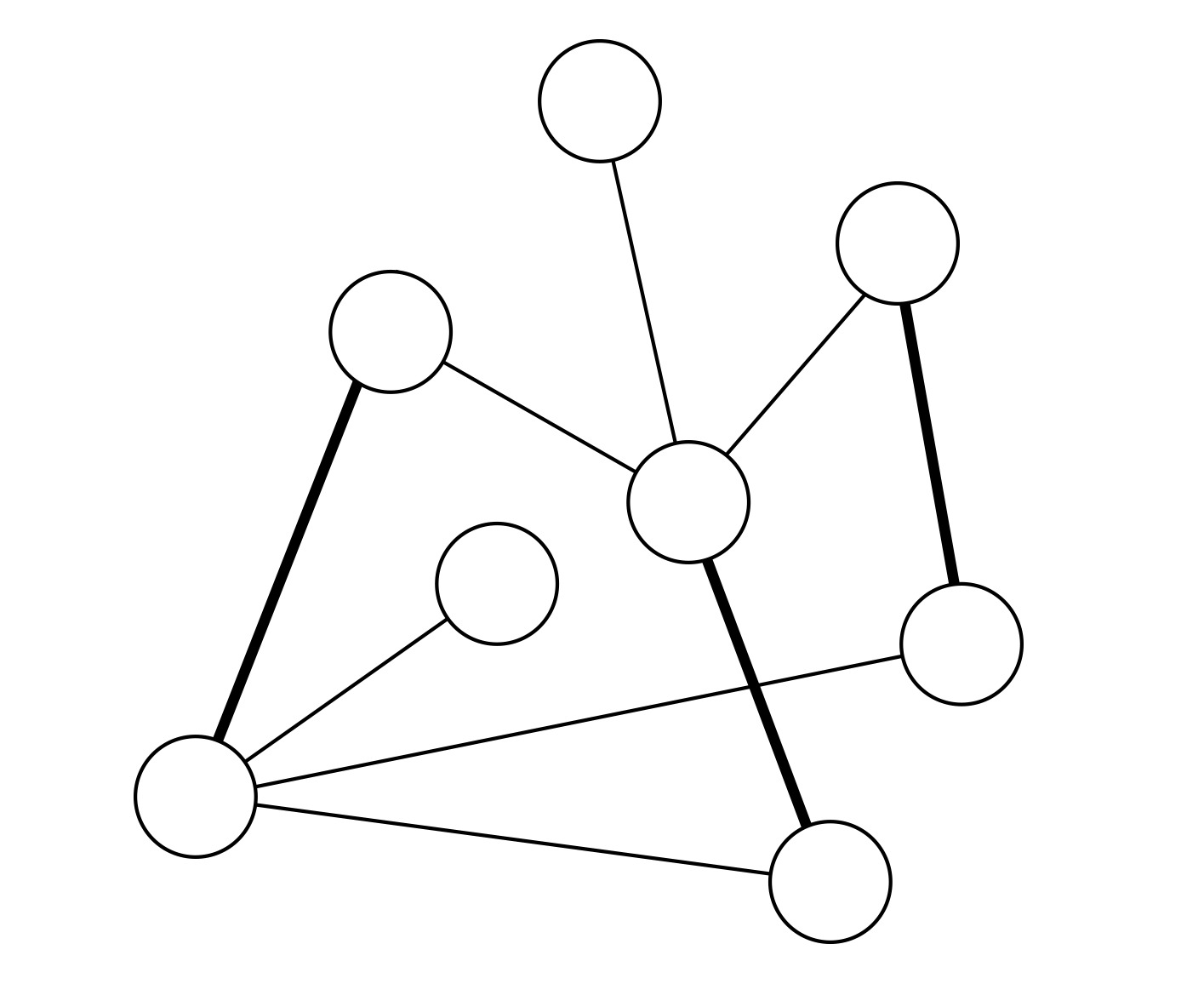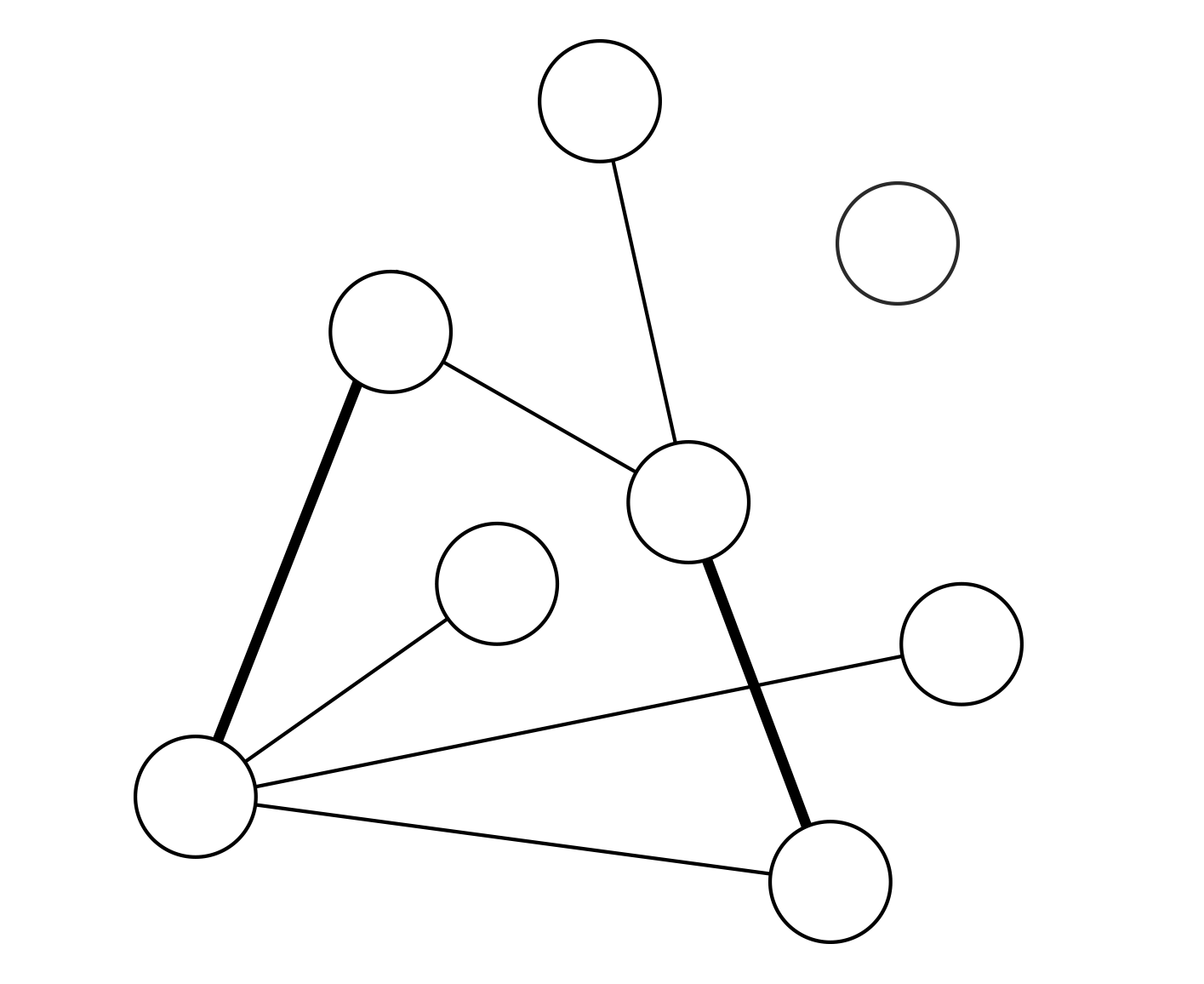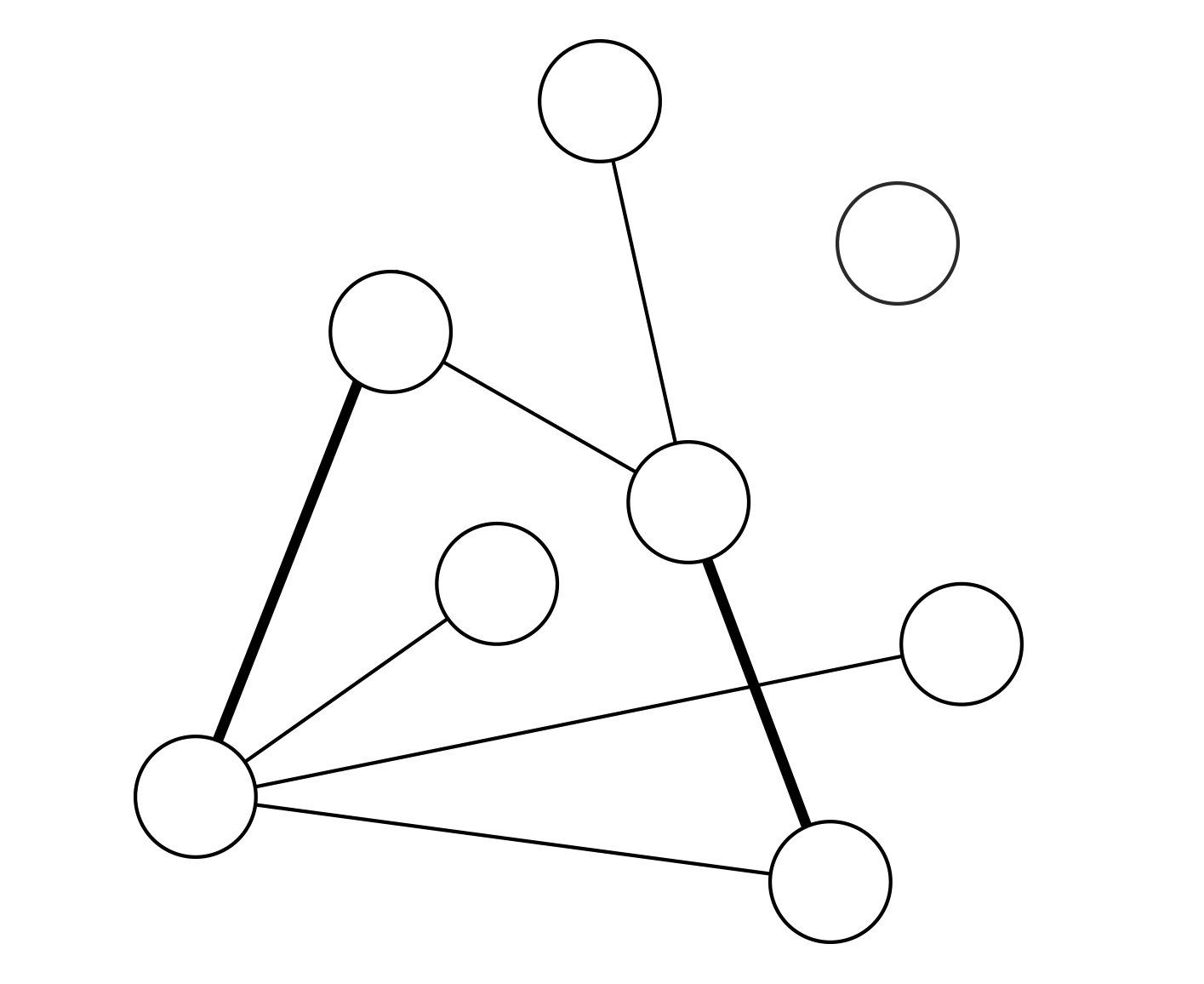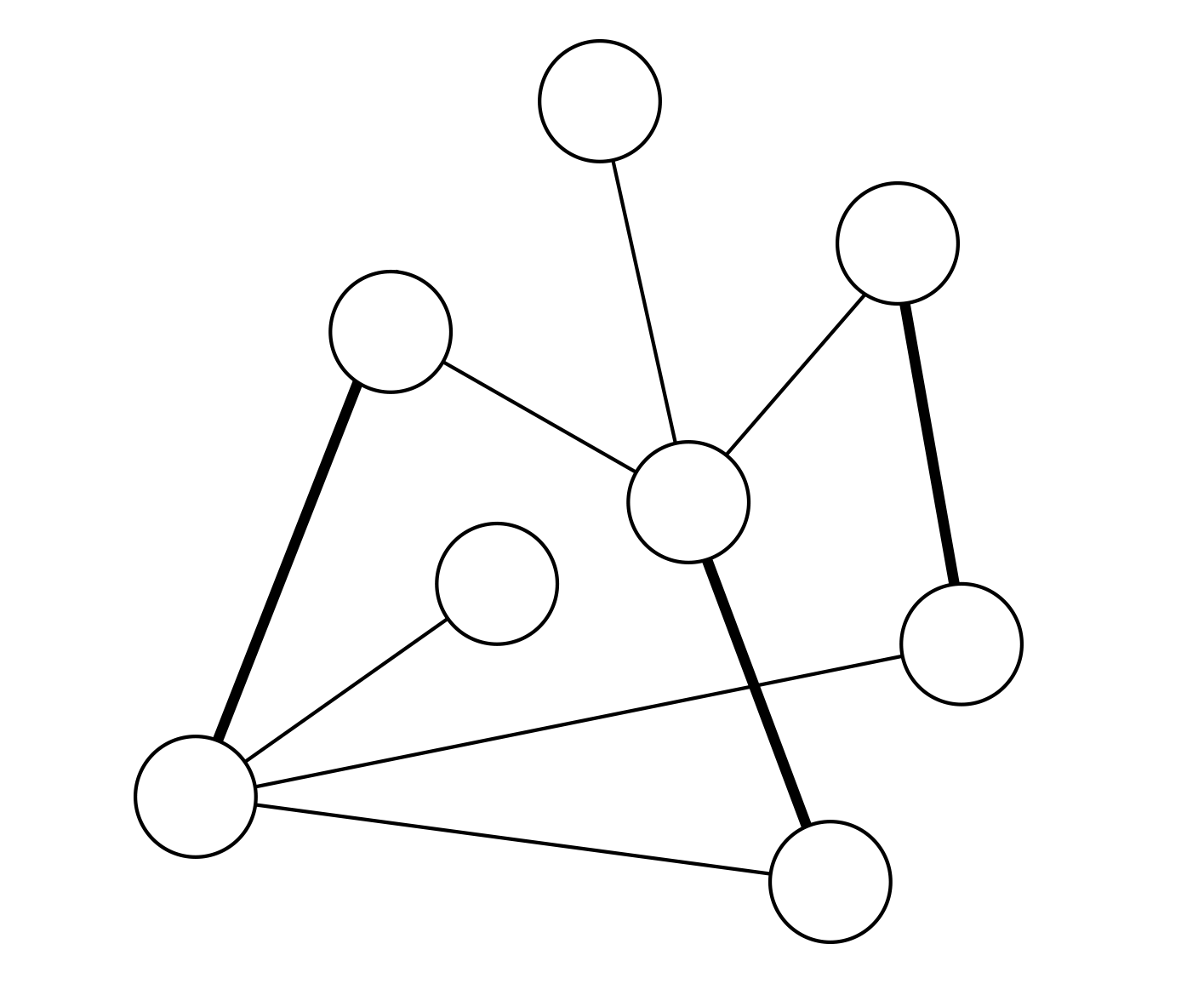
Recent studies show that in interdependent networks a very small failure in one network may lead to catastrophic consequences. Above a critical fraction of interdependent nodes, even a single node failure can invoke cascading failures that may abruptly fragment the system, whereas below this critical dependency a failure of a few nodes leads only to a small amount of damage to the system. So far, research has focused on interdependent random networks without space limitations. However, many real systems, such as power grids and the Internet, are not random but are spatially embedded. Here we analytically and numerically study the stability of interdependent spatially embedded networks modelled as lattice networks. Surprisingly, we find that in lattice systems, in contrast to non-embedded systems, there is no critical dependency and any small fraction of interdependent nodes leads to an abrupt collapse. We show that this extreme vulnerability of very weakly coupled lattices is a consequence of the critical exponent describing the percolation transition of a single lattice.
Personalized Medicine
Gestational Diabetes Mellitus (GDM) is one of the most common complications during pregnancy, defined as abnormal glucose regulation. One of the most common treatments for GDM patients is diet control of limited daily calorie intake to maintain blood glucose within the normal range and reduce the risk of complications. In recent years, the discovery of a link between disorders in the community structure of the gut microbiome and disease states of the host have led to ongoing efforts to develop microbiome-based therapies to treat these diseases. In the case of GDM, however, previous studies have found differences in only a few specific taxa but no community-level distinction between the structure of the microbiome in GDM patients and healthy patients. In addition, it is unknown whether diet control performed on GDM patients affects the community structure of their microbiome. Here, we propose a new method to analyze the microbial communities of GDM patients from the perspective of the networks of inter-species interactions in both group-level and personalized-level. The networks of the group of 30 healthy pregnant women (control group) exhibited a significant level of consistency over a period of two weeks. In the case of 27 GDM patients, after two weeks of calorie restriction therapy, the consistency of the networks was found to be lower than the control group. Furthermore, we found that the networks of the GDM patients became less similar to that of the control group after calorie restriction. In addition, analysis of the microbial networks of the individual GDM patients suggests an association between large deviations of the networks and abnormality of glucose regulation. These results are not captured by traditional community dissimilarity measures. Our findings demonstrate that microbial networks can provide new insights on the relation between the microbiome and GDM disease and diet control treatments. This approach may help us in developments of new diagnosis measures and individualized microbiome-based therapies.